Getting started¶
https://github.com/dask/dask-image/
conda¶
conda install -c conda-forge dask-imagepip¶
pip install dask-imageWhat's included?¶
- imread
- ndfilters
- ndfourier
- ndmorph
- ndmeasure
Function coverage¶

GPU support¶
Latest release includes GPU support for the modules:
- ndfilters
- imread
Still to do: ndfourier, ndmeasure, ndmorph*
*Done, pending cupy PR #3907
GPU benchmarking¶
| Architecture | Time |
|---|---|
| Single CPU Core | 2hr 39min |
| Forty CPU Cores | 11min 30s |
| One GPU | 1min 37s |
| Eight GPUs | 19s |
https://blog.dask.org/2019/01/03/dask-array-gpus-first-steps
Let's build a pipeline!¶
- Reading in data
- Filtering images
- Segmenting objects
- Morphological operations
- Measuring objects
%gui qt
We used image set BBBC039v1 Caicedo et al. 2018, available from the Broad Bioimage Benchmark Collection Ljosa et al., Nature Methods, 2012.
https://bbbc.broadinstitute.org/BBBC039
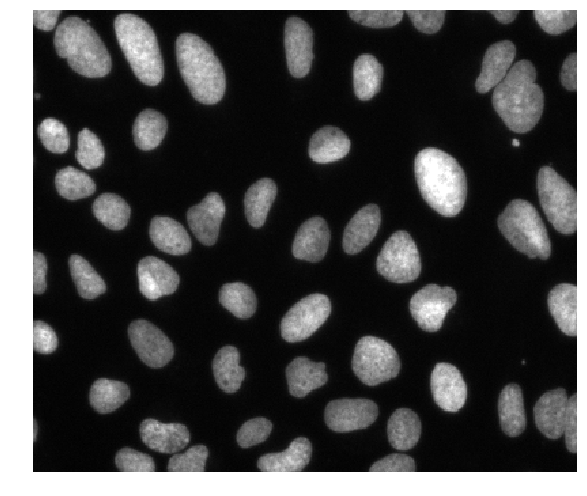
1. Reading in data¶
from dask_image.imread import imread
images = imread('data/BBBC039/images/*.tif')
# images_on_gpu = imread('data/BBBC039/images/*.tif', arraytype="cupy")
images
2. Filtering images¶
Denoising images with a small blur can improve segmentation later on.
from dask_image import ndfilters
smoothed = ndfilters.gaussian_filter(images, sigma=[0, 1, 1])
3. Segmenting objects¶
Pixels below the threshold value are background.
absolute_threshold = smoothed > 160
# Let's have a look at the images
import napari
from napari.utils import nbscreenshot
viewer = napari.Viewer()
viewer.add_image(absolute_threshold)
viewer.add_image(images, contrast_limits=[0, 2000])
nbscreenshot(viewer)
viewer.layers[-1].visible = False
napari.utils.nbscreenshot(viewer)
3. Segmenting objects (continued)¶
A better segmentation using local thresholding.
thresh = ndfilters.threshold_local(smoothed, images.chunksize)
threshold_images = smoothed > thresh
# Let's take a look at the images
viewer.add_image(threshold_images)
nbscreenshot(viewer)
4. Morphological operations¶
These are operations on the shape of a binary image.
Erosion¶

Dilation¶
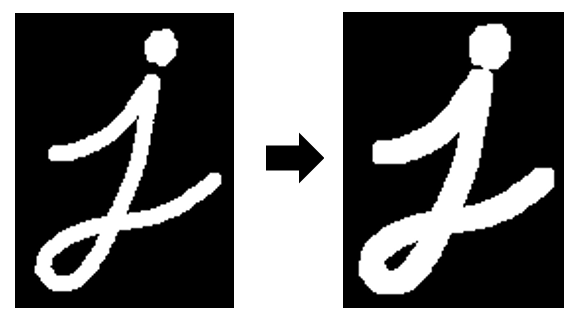
A morphological opening operation is an erosion, followed by a dilation.

from dask_image import ndmorph
import numpy as np
structuring_element = np.array([
[[0, 0, 0], [0, 0, 0], [0, 0, 0]],
[[0, 1, 0], [1, 1, 1], [0, 1, 0]],
[[0, 0, 0], [0, 0, 0], [0, 0, 0]]])
binary_images = ndmorph.binary_opening(threshold_images, structure=structuring_element)
5. Measuring objects¶
Each image has many individual nuclei, so for the sake of time we'll measure a small subset of the data.
from dask_image import ndmeasure
# Create labelled mask
label_images, num_features = ndmeasure.label(binary_images[:3], structuring_element)
index = np.arange(num_features - 1) + 1 # [1, 2, 3, ...num_features]
print("Number of nuclei:", num_features.compute())
# Let's look at the labels
viewer.add_labels(label_images)
viewer.dims.set_point(0, 0)
nbscreenshot(viewer)
Measuring objects (continued)¶
# Measure objects in images
area = ndmeasure.area(images[:3], label_images, index)
mean_intensity = ndmeasure.mean(images[:3], label_images, index)
# Run computation and plot results
import matplotlib.pyplot as plt
plt.scatter(area, mean_intensity, alpha=0.5)
plt.gca().update(dict(title="Area vs mean intensity", xlabel='Area (pixels)', ylabel='Mean intensity'))
plt.show()
viewer.close() # close the napari viewer
The full pipeline¶
import numpy as np
from dask_image.imread import imread
from dask_image import ndfilters, ndmorph, ndmeasure
images = imread('data/BBBC039/images/*.tif')
smoothed = ndfilters.gaussian_filter(images, sigma=[0, 1, 1])
thresh = ndfilters.threshold_local(smoothed, blocksize=images.chunksize)
threshold_images = smoothed > thresh
structuring_element = np.array([[[0, 0, 0], [0, 0, 0], [0, 0, 0]], [[0, 1, 0], [1, 1, 1], [0, 1, 0]], [[0, 0, 0], [0, 0, 0], [0, 0, 0]]])
binary_images = ndmorph.binary_closing(threshold_image)
label_images, num_features = ndmeasure.label(binary_image)
index = np.arange(num_features)
area = ndmeasure.area(images, label_images, index)
mean_intensity = ndmeasure.mean(images, label_images, index)
Custom functions¶
What if you want to do something that isn't included?
- scikit-image apply_parallel()
- dask map_overlap / map_blocks
- dask delayed
Scaling up computation¶
Use dask-distributed to scale up from a laptop onto a supercomputing cluster.
from dask.distributed import Client
# Setup a local cluster
# By default this sets up 1 worker per core
client = Client()
client.cluster
dask-image¶
- Install:
condaorpip install dask-image - Documentation: https://dask-image.readthedocs.io
- GitHub: https://github.com/dask/dask-image/
# CPU example
import numpy as np
import dask.array as da
from dask_image.ndfilters import convolve
s = (10, 10)
a = da.from_array(np.arange(int(np.prod(s))).reshape(s), chunks=5)
w = np.ones(a.ndim * (3,), dtype=np.float32)
result = convolve(a, w)
result.compute()
# Same example moved to the GPU
import cupy # <- import cupy instead of numpy (version >=7.7.0)
import dask.array as da
from dask_image.ndfilters import convolve
s = (10, 10)
a = da.from_array(cupy.arange(int(cupy.prod(cupy.array(s)))).reshape(s), chunks=5) # <- cupy dask array
w = cupy.ones(a.ndim * (3,)) # <- cupy dask array
result = convolve(a, w)
result.compute()